
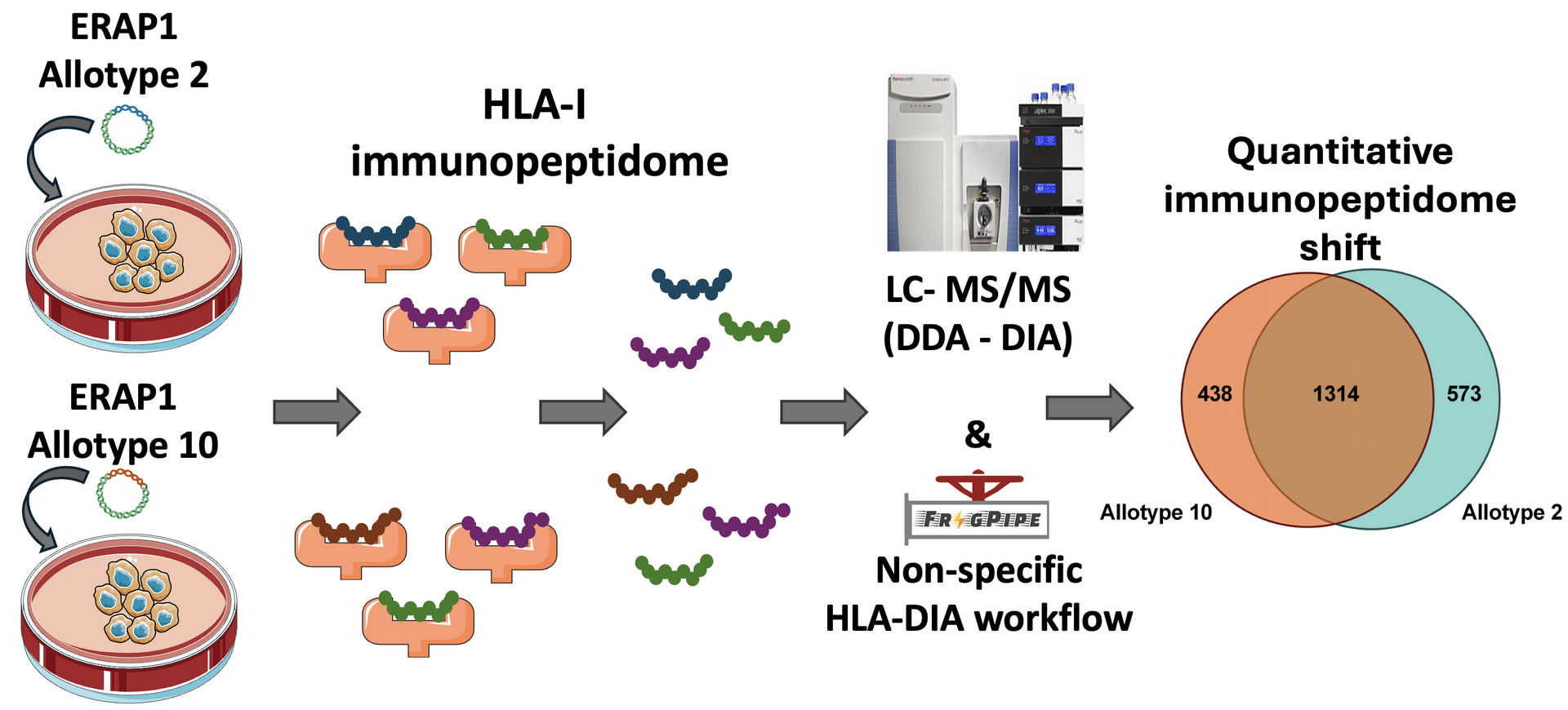
Endoplasmic reticulum aminopeptidase 1 (ERAP1) is a polymorphic enzyme that shapes the peptide repertoire presented by MHC class I molecules and can regulate adaptive immune responses in cancer and autoimmunity. Common missense polymorphisms in ERAP1 modulate its activity and are found in specific allotypes in humans. ERAP1 allotypes are linked to predisposition to HLA-associated inflammatory diseases such as psoriasis and Behçet's disease, through the generation of specific CD8+ T cell populations targeting disease-specific HLAs. Given the established broad effects of ERAP1 activity on the cellular immunopeptidome, we hypothesised that ERAP1 allotypic variation may lead to broad immunopeptidome shifts capable of triggering the observed antigenic responses. To test this hypothesis, we generated two A375 melanoma cell lines, each one expressing one of the most common, disease-associated ERAP1 allotypes, namely allotypes 2 or 10. Comparison of the immunopeptidome of these two cell lines showed only minor differences in peptide sequences presented but extensive changes in abundance that included alterations in length distribution, binding affinity, and sequence motifs. Our results suggest that enzymatic differences between ERAP1 allotypes are reflected primarily in the quantitative composition of the cellular immunopeptidome. These quantitative changes may constitute a mechanism that underlies ERAP1-allotypic associations with HLA-associated autoimmunity and variable immune responses (Immunology 2025)
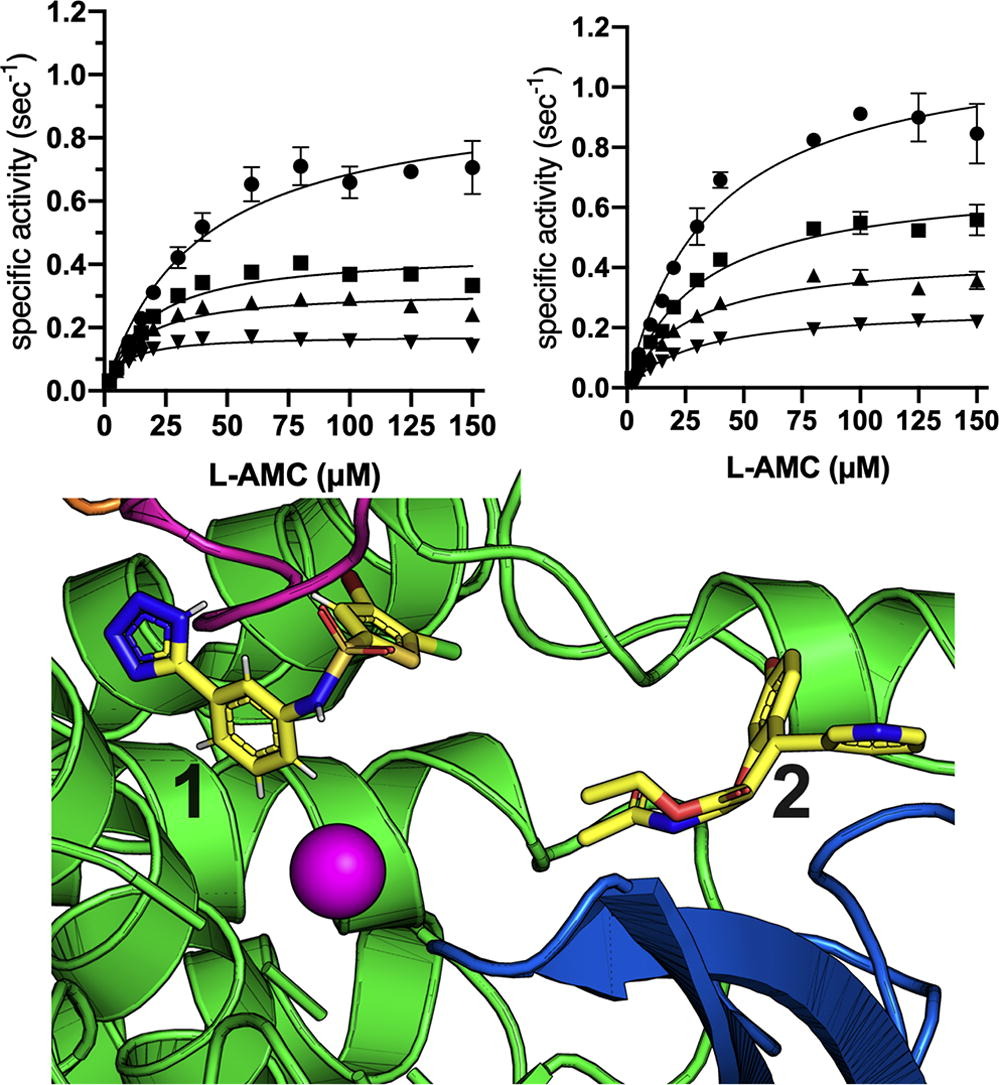
Inhibition of Insulin-Regulated Aminopeptidase is being actively explored for the treatment of several human diseases and several classes of inhibitors have been developed although no clinical applications have been reported yet. Here, we combine enzymological analysis with x-ray crystallography to investigate the mechanism employed by two of the most studied inhibitors of IRAP, an aryl sulfonamide and a 2-amino-4H-benzopyran named HFI-419. Although both compounds have been hypothesized to target the enzyme's active site by competitive mechanisms, we discovered that they instead target previously unidentified proximal allosteric sites and utilize non-competitive inhibition mechanisms. X-ray crystallographic analysis demonstrated that the aryl sulfonamide stabilizes the closed, more active, conformation of the enzyme whereas HFI-419 locks the enzyme in a semi-open, and likely less active, conformation. HFI-419 potency is substrate-dependent and fails to effectively block the degradation of the physiological substrate cyclic peptide oxytocin. Our findings demonstrate alternative mechanisms for inhibiting IRAP through allosteric sites and conformational restricting and suggest that the pharmacology of HFI-419 may be more complicated than initially considered. Such conformation-specific interactions between IRAP and small molecules can be exploited for the design of more effective second-generation allosteric inhibitors (JMB 2024).
 ER aminopeptidase 1 (ERAP1) is an ER-resident aminopeptidase that excises N-terminal residues of peptides that then bind onto Major Histocompatibility Complex I molecules (MHC-I) and indirectly modulates adaptive immune responses. ERAP1 contains an allosteric regulatory site that accommodates the C-terminus of at least some peptide substrates, raising questions about its exact influence on antigen presentation and the potential of allosteric inhibition for cancer immunotherapy. We used an inhibitor that targets this regulatory site to study its effect on the immunopeptidome of a human cancer cell line. The immunopeptidomes of allosterically inhibited and ERAP1 KO cells contain high-affinity peptides with sequence motifs consistent with the cellular HLA class I haplotypes but are strikingly different in peptide composition. Compared to KO cells, allosteric inhibition did not affect the length distribution of peptides and skewed the peptide repertoire both in terms of sequence motifs and HLA allele utilization, indicating significant mechanistic differences between the two ways of disrupting ERAP1 function. These findings suggest that the regulatory site of ERAP1 plays distinct roles in antigenic peptide selection, which should be taken into consideration when designing therapeutic interventions targeting the cancer immunopeptidome (
Eur J Immunol 2023).
ER aminopeptidase 1 (ERAP1) is an ER-resident aminopeptidase that excises N-terminal residues of peptides that then bind onto Major Histocompatibility Complex I molecules (MHC-I) and indirectly modulates adaptive immune responses. ERAP1 contains an allosteric regulatory site that accommodates the C-terminus of at least some peptide substrates, raising questions about its exact influence on antigen presentation and the potential of allosteric inhibition for cancer immunotherapy. We used an inhibitor that targets this regulatory site to study its effect on the immunopeptidome of a human cancer cell line. The immunopeptidomes of allosterically inhibited and ERAP1 KO cells contain high-affinity peptides with sequence motifs consistent with the cellular HLA class I haplotypes but are strikingly different in peptide composition. Compared to KO cells, allosteric inhibition did not affect the length distribution of peptides and skewed the peptide repertoire both in terms of sequence motifs and HLA allele utilization, indicating significant mechanistic differences between the two ways of disrupting ERAP1 function. These findings suggest that the regulatory site of ERAP1 plays distinct roles in antigenic peptide selection, which should be taken into consideration when designing therapeutic interventions targeting the cancer immunopeptidome (
Eur J Immunol 2023).
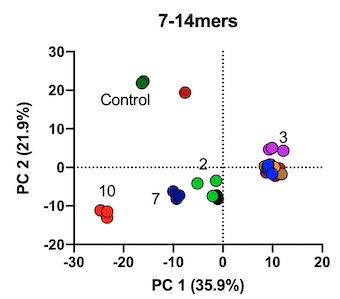 Population genetic variability in immune system genes can often underlie variability in immune responses to pathogens. Cytotoxic T-lymphocytes are emerging as critical determinants of both SARS-CoV-2 infection severity and long-term immunity, after either recovery or vaccination. A hallmark of COVID-19 is its highly variable severity and breadth of immune responses between individuals. To address the underlying mechanisms behind this phenomenon we analyzed the proteolytic processing of S1 spike glycoprotein precursor antigenic peptides across 10 common allotypes of ER aminopeptidase 1 (ERAP1), a polymorphic intracellular enzyme that can regulate cytotoxic T-lymphocyte responses by generating or destroying antigenic peptides. We utilized a systematic proteomic approach that allows the concurrent analysis of hundreds of trimming reactions in parallel, thus better emulating antigen processing in the cell. While all ERAP1 allotypes were capable of producing optimal ligands for MHC class I molecules, including known SARS-CoV-2 epitopes, they presented significant differences in peptide sequences produced, suggesting allotype-dependent sequence biases. Allotype 10, previously suggested to be enzymatically deficient, was rather found to be functionally distinct from other allotypes. Our findings suggest that common ERAP1 allotypes can be a major source of heterogeneity in antigen processing and through this mechanism contribute to variable immune responses in COVID-19. (J Biol Chem. 2021).
Population genetic variability in immune system genes can often underlie variability in immune responses to pathogens. Cytotoxic T-lymphocytes are emerging as critical determinants of both SARS-CoV-2 infection severity and long-term immunity, after either recovery or vaccination. A hallmark of COVID-19 is its highly variable severity and breadth of immune responses between individuals. To address the underlying mechanisms behind this phenomenon we analyzed the proteolytic processing of S1 spike glycoprotein precursor antigenic peptides across 10 common allotypes of ER aminopeptidase 1 (ERAP1), a polymorphic intracellular enzyme that can regulate cytotoxic T-lymphocyte responses by generating or destroying antigenic peptides. We utilized a systematic proteomic approach that allows the concurrent analysis of hundreds of trimming reactions in parallel, thus better emulating antigen processing in the cell. While all ERAP1 allotypes were capable of producing optimal ligands for MHC class I molecules, including known SARS-CoV-2 epitopes, they presented significant differences in peptide sequences produced, suggesting allotype-dependent sequence biases. Allotype 10, previously suggested to be enzymatically deficient, was rather found to be functionally distinct from other allotypes. Our findings suggest that common ERAP1 allotypes can be a major source of heterogeneity in antigen processing and through this mechanism contribute to variable immune responses in COVID-19. (J Biol Chem. 2021).
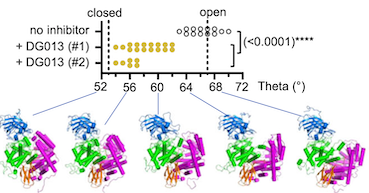 The endoplasmic-reticulum aminopeptidase ERAP1 processes antigenic peptides for loading on MHC-I proteins and recognition by CD8 T cells as they survey the body for infection and malignancy. Crystal structures have revealed ERAP1 in either open or closed conformations, but whether these occur in solution and are involved in catalysis is not clear. Here, we assess ERAP1 conformational states in solution in the presence of substrates, allosteric activators, and inhibitors by small-angle X-ray scattering. We also characterize changes in protein conformation by X-ray crystallography, and we localize alternate C-terminal binding sites by chemical crosslinking. Structural and enzymatic data suggest that the structural reconfigurations of ERAP1 active site are physically linked to domain closure and are promoted by binding of long peptide substrates. These results clarify steps required for ERAP1 catalysis, demonstrate the importance of conformational dynamics within the catalytic cycle, and provide a mechanism for the observed allosteric regulation and Lys/Arg528 polymorphism disease association. (Nature Communications 2021).
The endoplasmic-reticulum aminopeptidase ERAP1 processes antigenic peptides for loading on MHC-I proteins and recognition by CD8 T cells as they survey the body for infection and malignancy. Crystal structures have revealed ERAP1 in either open or closed conformations, but whether these occur in solution and are involved in catalysis is not clear. Here, we assess ERAP1 conformational states in solution in the presence of substrates, allosteric activators, and inhibitors by small-angle X-ray scattering. We also characterize changes in protein conformation by X-ray crystallography, and we localize alternate C-terminal binding sites by chemical crosslinking. Structural and enzymatic data suggest that the structural reconfigurations of ERAP1 active site are physically linked to domain closure and are promoted by binding of long peptide substrates. These results clarify steps required for ERAP1 catalysis, demonstrate the importance of conformational dynamics within the catalytic cycle, and provide a mechanism for the observed allosteric regulation and Lys/Arg528 polymorphism disease association. (Nature Communications 2021).
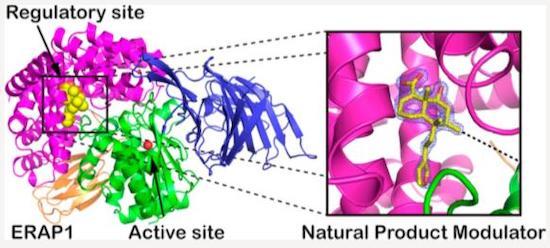 ER aminopeptidase 1 (ERAP1) is an intracellular enzyme that generates antigenic peptides and is an emerging target for cancer immunotherapy and the control of autoimmunity. ERAP1 inhibitors described previously target the active site and are limited in selectivity, minimizing their clinical potential. To address this, we targeted the regulatory site of ERAP1 using a high- throughput screen and discovered a small molecule hit that is highly selective for ERAP1. (4aR,5S,6R,8S,8aR)-5-(2-(Furan-3- yl)ethyl)-8-hydroxy-5,6,8a-trimethyl-3,4,4a,5,6,7,8,8a-octahydronaphthalene-1-carboxylic acid is a natural product found in Dodonaea viscosa that constitutes a submicromolar, highly selective, and cell-active modulator of ERAP1. Although the compound activates hydrolysis of small model substrates, it is a competitive inhibitor for physiologically relevant longer peptides. Crystallographic analysis confirmed that the compound targets the regulatory site of the enzyme that normally binds the C-terminus of the peptide substrate. Our findings constitute a novel starting point for the development of selective ERAP1 modulators that have potential for further clinical development. (Journal of Medicinal Chemistry 2020)
ER aminopeptidase 1 (ERAP1) is an intracellular enzyme that generates antigenic peptides and is an emerging target for cancer immunotherapy and the control of autoimmunity. ERAP1 inhibitors described previously target the active site and are limited in selectivity, minimizing their clinical potential. To address this, we targeted the regulatory site of ERAP1 using a high- throughput screen and discovered a small molecule hit that is highly selective for ERAP1. (4aR,5S,6R,8S,8aR)-5-(2-(Furan-3- yl)ethyl)-8-hydroxy-5,6,8a-trimethyl-3,4,4a,5,6,7,8,8a-octahydronaphthalene-1-carboxylic acid is a natural product found in Dodonaea viscosa that constitutes a submicromolar, highly selective, and cell-active modulator of ERAP1. Although the compound activates hydrolysis of small model substrates, it is a competitive inhibitor for physiologically relevant longer peptides. Crystallographic analysis confirmed that the compound targets the regulatory site of the enzyme that normally binds the C-terminus of the peptide substrate. Our findings constitute a novel starting point for the development of selective ERAP1 modulators that have potential for further clinical development. (Journal of Medicinal Chemistry 2020)